GMS:Null Space Monte Carlo
The Null Space Monte Carlo (NSMC) method is used to create multiple calibrated MODFLOW models. The method is selected in the Stochastic Options dialog, however, the option requires that the model has already been calibrated using PEST SVD-Assist.
There is always a significant amount of uncertainty associated with a groundwater model. This uncertainty can be associated with the conceptual model or the field data or the input parameters of the model. Some model parameters, such as hydraulic conductivity, are particularly prone to uncertainty. This uncertainty can be reduced by calibrating a model to observation data (monitoring wells, stream flows, etc.). However, even a well-calibrated model can have significant uncertainty associated with it.
The challenge with a groundwater model is to explore the uncertainty while maintaining a calibrated model (good fit with field measured values). The generation of model input parameters that will respect calibration constraints is a time consuming matter since a parameter estimation exercise must be undertaken for each new set of parameters. However, two tools provided with PEST greatly reduce the amount of time required to perform calibration-constrained Monte Carlo analysis of a groundwater model. The first is the use of SVD-Assist in the calibration process. The second is the pre-calibration null space[1]projection of differences between the stochastic parameter fields and the simplified “calibration parameter field” that is decreed to “calibrate the model”. By using both of these utilities it is possible to obtain parameter sets that respect both the stochastic variability of the subsurface as well as the field measured values; this is accomplished with only a handful of runs per realization.
NSMC is a method for quantifying the amount of uncertainty associated with the prediction. Instead of creating a single calibrated model, PEST NSMC is used to create multiple calibrated models. The model can be modified to account for future conditions, then multiple models can be run using the different parameter values that PEST calculated. A distribution of results for this prediction can then be viewed.
There are many steps involved in running PEST with the Null Space Monte Carlo method. GMS runs several PEST utilities to make all of this happen. The following is a list of the steps involved:
- PEST runs MODFLOW once for each parameter to compute a Jacobian matrix. In this case, there is a steady state model and Parallel PEST is being used, so this step is completed fairly quickly.
- PPCOV is a PEST utility that generates uncertainty information for the pilot points. The PPCOV utility generates a covariance matrix for the pilot points. This matrix is created using the variogram that is specified as part of the interpolation options for the pilot points. For this reason, kriging must be used with the pilot points if using the Null Space Monte Carlo method.
- RANDPAR is a PEST utility that generates random values for the parameters that are being randomized, saving one file for each realization.
- PNULPAR is a PEST utility that modifies the random parameter values through Null Space projection, generating one file for each realization. An explanation of how PNULPAR calculates values is given below:
In this process the difference is first taken between each parameter set and the calibration parameter; this difference is then projected onto the calibration null space, and the projected difference re-added to the calibration parameter set. If the model were linear, and if the cutoff between null and solution spaces were sharp, each new parameter set obtained in this way would calibrate the model with an identical objective function to that achieved using the calibration parameter set. However, in general, random parameter sets modified in this manner do not result in perfectly calibrated models, and will thus require adjustment in order that calibration constraints be respected. Such adjustment takes place through alterations made to the calibration solution space (leaving the null space untouched).
- SVDAPREP is a PEST utility that creates a new PEST input file (PST) using information from the original PEST run in step 1. PEST then runs SVD-assisted parameter estimation, greatly reducing the number of model runs required for each PEST iteration.
- For each realization, two steps are required:
- PARREP is a PEST utility that substitutes the parameter values generated by PNULPAR in step 4 for the current realization.
- PEST is launched using the parameter values for the current realization and runs until the objective function is at or below PHISTOPTHRESH or until that maximum number of PEST iterations is reached.
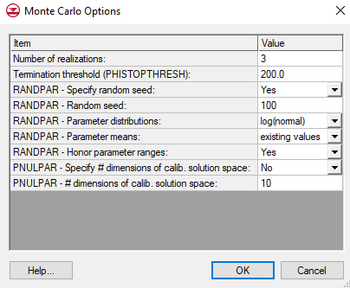
Monte Carlo Options
The Monte Carlo Options dialog is reached through the Stochastic Options dialog by selecting the Options button available when the PEST NSMC option is selected. The dialog has the following options:
- Number of realizations: Enter a positive value.
- Termination threshold (PHISTOPTHRESH): This value tells PEST to stop when the total model error is the specified value or less.
- RANDPAR – Specify random seed: Set to "Yes" to change the random seed defaults. Set to "No" to keep default values.
- RANDPAR – Random seed: Enter a postive integer specified to allow replication of the results of the sample solution. Random number sequences are identical for the same random number seed.
- RANDPAR – Parameter distributions: Choose between two probability distributions for random number generation.
- "log(normal)": Requires parameter variances/covariances to be supplied.
- "log(uniform)": In this case the upper and lower end of the parameter range is taken as the upper and lower bounds of respective parameters as recorded in the PEST control file. If a parameter is log-transformed, then the log of the parameter is assumed to possess a uniform distribution rather than the parameter itself. No correlation is permitted between parameter values if a uniform distribution is specified.
- RANDPAR – Parameter means: In generating random numbers which belong to a normal distribution, a mean value is required.
- "existing values": Accept the initial parameter value set in the control file.
- "range midpoints": Compute the mean as midway between the lower and upper bound of each parameter set in the control file.
- RANDPAR – Honor parameter ranges: In preserving the ratios between tied and parent parameter values, the former may transgress their bounds as the latter are assigned random values between those bounds. Can be set to "No" or "Yes".
- “No”: Ignores the fact that tied parameters may transgress their bounds.
- “Yes”: The tied parameters are clipped at their bounds, thereby destroying the tied parameter.
- PNULPAR – Specify # dimensions of calib. solution space: Enter "Yes" or "No".
- "No": Will not constrain calibration.
- "Yes": Will use calibration constraint entered in the next field.
- PNULPAR – # dimension of calib. solution space: Enter a positive integer to constrain the calibration. See PEST manual for more information.
Related Topics
| [hide] GMS – Groundwater Modeling System | ||
|---|---|---|
| Modules: | 2D Grid • 2D Mesh • 2D Scatter Point • 3D Grid • 3D Mesh • 3D Scatter Point • Boreholes • GIS • Map • Solid • TINs • UGrids | |
| Models: | FEFLOW • FEMWATER • HydroGeoSphere • MODAEM • MODFLOW • MODPATH • mod-PATH3DU • MT3DMS • MT3D-USGS • PEST • PHT3D • RT3D • SEAM3D • SEAWAT • SEEP2D • T-PROGS • ZONEBUDGET | |
| Aquaveo | ||