WMS:GSSHA Automated Calibration: Difference between revisions
No edit summary |
|||
| (70 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{TOCright}} | {{TOCright}} | ||
The GSSHA model allows for both automated and [[WMS:GSSHA Manual Calibration|manual]] calibration. There is a [[WMS:WMS_Tutorials|WMS tutorial]] that describes how to set up a basic calibration model. | |||
GSSHA has a PEST-based automated calibration routine, as described on the [http://gsshawiki.com/Alternate_Run_Modes:ERDC_Automated_Model_Calibration_Software GSSHA wiki]. WMS writes the parameters, observed data, and calibration files necessary to run GSSHA in any of the supported GSSHA calibration modes. Enter in the needed data by turning on ''Calibrate'' in the list on the right in the [[WMS:GSSHA Job Control|''Job Control'' dialog]]. | |||
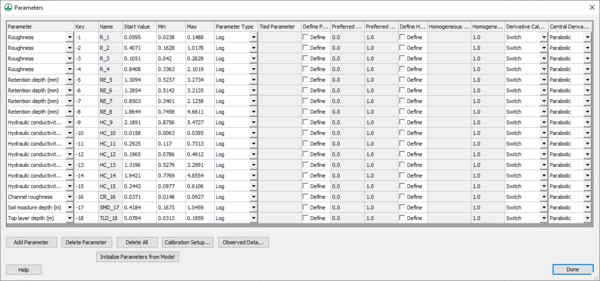
Clicking on the '''Edit parameters''' button brings up the ''Parameters'' dialog (below) where parameters can be selected to be calibrated and edited, including the maximum, minimum, and initial values for each parameter. | |||
[[ | [[File:CalibParams.png|thumb|none|left|600 px|The ''Parameters'' dialog for auto calibration.]] | ||
==Defining Key Values== | |||
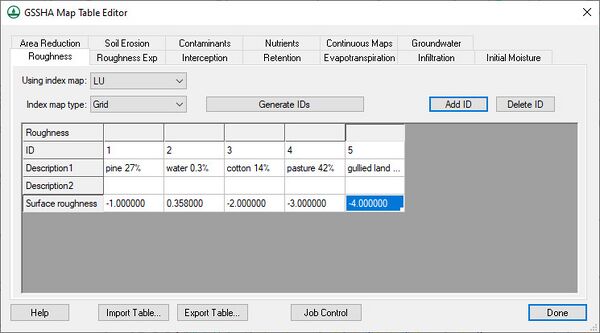
Select adjustable parameters by defining ''key'' values as negative integers in the regular GSSHA interface and then assigning starting, minimum, and maximum values to each of these parameters in the ''Parameters'' dialog. For example, to set overland Manning’s roughness values as an adjustable model parameter, specify negative integers for the values in the mapping table dialog as shown below: | |||
[[File:MappingCalibParams.jpg|thumb|none|left|600 px|''GSHHA Map Table Editor'' dialog with ''Surface roughness'' row showing negative integers.]] | |||
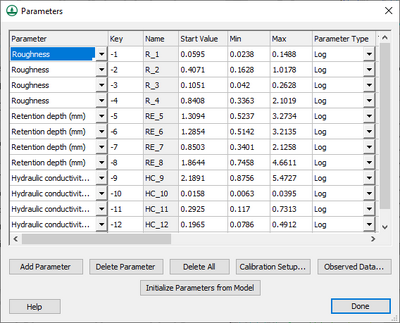
The | Then define the key values in the calibration ''Parameters'' dialog. The '''Initialize Parameters from Model''' button makes defining key values easy. This button searches the model for any parameters that are defined as negative integers and assigns these parameters to the dialog. Starting, minimum, and maximum values should be defined for each key value for the calibration engine. Other parameter information should also be defined, including: | ||
*Whether parameters are fixed or tied to other parameters | |||
*The methods used to calculate derivatives for each parameter | |||
*Defining regularization parameters for prior calibration run information | |||
*Defining preferred values (where a specific value is set as an optimal value for the calibration parameter) or homogeneous values (where one or more calibration parameters are linked to another calibration parameter in an attempt to get these values as close as possible). | |||
[[File:ParameterKeyValues.png|thumb|none|left|400px|''Parameters'' dialog with key values entered in ''Start Value'', ''Min'', and ''Max'' columns.]] | |||
[[Image: | ==Defining Observed Data== | ||
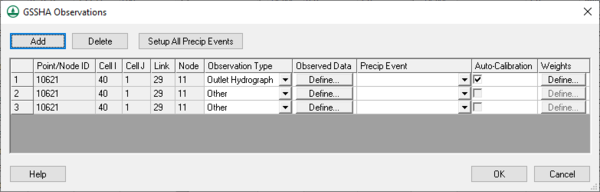
[[Image:CalibrationObserved.png|thumb|right|600 px|Calibration ''GSSHA Observations'' dialog.]] | |||
Clicking the '''Observed Data...''' button in the ''Calibration Parameters'' dialog opens the [[WMS:GSSHA Observations|''GSSHA Observations'' dialog]] (right). In this dialog, enter in various types of observed data (including time series hydrograph data) for each event of the simulation. It is not necessary to associate a rainfall event with each observed data time series. | |||
This dialog shows all the feature points with observed data and allows turning automated calibration on or off for each of these observations. All automated calibration methods in WMS support automated calibration of the following data types at any computation point in the watershed model: | |||
* Overland Depth | |||
* Infiltration Depth | |||
* Surface Moisture | |||
* Grid Suspended Sediment (TSS) Concentration | |||
* Channel Depth | |||
* Channel Flow | |||
* Channel Total Suspended Sediment (TSS) Concentration | |||
* Groundwater Head | |||
* Outlet Hydrograph (only at the watershed outlet point) | |||
* Snow Water Equivalent | |||
* Tile Drain Discharge (in a GSSHA Storm Drain coverage) | |||
[[ | The [[WMS:GSSHA Observations|''GSSHA Observations'' dialog]] can be accessed from the feature point/node attribute dialog in the GSSHA and the GSSHA Storm Drain coverages. If WMS is running the default LM/SLM-based calibration, associate weights with each observation value in the XY series by clicking on the '''Weights''' button in the observation window and defining weights for each value. WMS defaults all the weights for observed data points to 1.0, but this value could be modified to give higher weights to certain observed values. | ||
==Defining Calibration Setup Parameters== | |||
[[Image:CalibrationParams.png|thumb|right|225 px|''Calibration Setup'' dialog.]] | |||
The ''' | The calibration setup parameters are accessed by clicking the '''Calibration Setup...''' button to bring up the ''Calibration Setup'' dialog: | ||
The parameters in this dialog are used for creating the calibration control file. This file, along with the parameters and observed data files are written out by WMS when saving a GSSHA project. | |||
The following options are in this dialog: | |||
*''Calibration method'' – A drop-down with a list of calibration methods. The [http://gsshawiki.com/Alternate_Run_Modes:Alternate_Run_Modes GSSHA wiki] has more information on these methods. | |||
**"Levenberg-Marquardt (LM)/Secant LM (SLM)" – Uses local search to optimize model parameters. | |||
**"Multistart (MS)" – Uses global search to optimize model parameters. | |||
**"Trajectory Repulsion (TR)" – Uses global search to optimize model parameters. | |||
**"Multilevel Single Linkage (MLSL)" – Uses global search to optimize model parameters.<!-- | |||
**"Shuffled Complex Evolution (SCE)" – Uses global search to optimize model parameters. As this method is now deprecated in GSSHA, it is not recommended this be selected. --> | |||
*''LM/SLM parameters'' | |||
**''Run Secant LM (SLM) method'' – Turning on this option to run the SLM method sets the input file flag to run the SLM method instead of the LM (Levenberg-Marquardt) local search method. The SLM method is an efficiency enhancement to the LM method and is the default local search optimization method for a GSSHA calibration in the WMS interface. | |||
***''Update Jacobian matrix at this ratio'' – Allows adjusting of the Jacobian matrix update ratio. | |||
**''Update Jacobian columns 0 to'' – Change the number of Jacobian columns. | |||
**''Use Tikhonov regularization'' – Turns Tikhonov regularization off or on. Tikhonov regularization provides a way to adjust the balance between fitting the solution to the observed data and fitting to the regularization relationships by defining a regularization weight factor and running Tikhonov regularization using this weight factor. The [http://gsshawiki.com/Alternate_Run_Modes:Efficient_Local_Search GSSHA wiki] has more information. | |||
**'''Advanced LM/SLM Parameters...''' – Brings up the [[#Advanced LM/SLM Parameters dialog|''Advanced LM/SLM Parameters'' dialog]]. | |||
*''Multistart parameters'' | |||
**'''Edit MS Parameters...''' – Brings up the [[#Multistart Parameters dialog|''Multistart Parameters'' dialog]]. | |||
*''Trajectory repulsion (TR) parameters'' | |||
**'''Edit TR Parameters...''' – Brings up the [[#Trajectory Repulsion (TR) Parameters dialog|''Trajectory Repulsion (TR) Parameters'' dialog]]. | |||
*''Multilevel single linkage (MLSL) parameters'' | |||
**'''Edit MLSL Parameters...''' – Brings up the [[#Multilevel Single Linkage (MLSL) Parameters dialog|''Multilevel Single Linkage (MLSL) Parameters'' dialog]]. | |||
*''SCE parameters'' section | |||
**SCE is deprecated in GSSHA and is not supported beginning in WMS 10.0. | |||
==Advanced LM/SLM Parameters dialog== | |||
[[Image:AdvancedLmSlmParameters.png|thumb|right|300 px|''Advanced LM/SLM Parameters'' dialog]] | |||
The ''Advanced LM/SLM Parameters'' dialog can be used to set advanced calibration settings when running an LM/SLM calibration. Some of the descriptions below were taken from the 2013 PEST User Manual. The following options are available in the ''Advanced LM/SLM Parameters'' section: | |||
*''Estimate parameter sensitivity'' | |||
*:Turn this option on to write a "-1" to the NOPTMAX parameter in the PEST control file. This tells PEST to only run a sensitivity analysis instead of a full parameter optimization run. The sensitivity file is imported into WMS if reading the calibration solution. | |||
*''Max # of optimization iterations'' (NOPTMAX) | |||
*:A value of "20" to "30" is often appropriate. Sets the maximum number of optimization iterations that PEST is permitted to undertake on a particular parameter estimation run. To trigger PEST termination by other criteria more indicative of parameter convergence to an optimal set or of the futility of further processing, set this variable very high. | |||
*:If NOPTMAX is set to zero, PEST will not calculate the Jacobian matrix. Instead, it will terminate execution after just one model run. This setting can thus be used when calculating the objective function corresponding to a particular parameter set and/or to inspect observation residuals corresponding to that parameter set. | |||
*''Initial Marquardt Lambda value'' (RLAMBDA1) | |||
*:This real variable is the initial Marquardt lambda. An initial value of "1.0" to "10.0" is appropriate for most models. PEST attempts parameter improvement using a number of different Marquardt lambdas during any one optimization iteration. However, in the course of the overall parameter estimation process, the Marquardt lambda generally gets smaller. | |||
*:Provide a higher initial Marquardt lambda if PEST complains that the normal matrix is not positive definite. For high values of the Marquardt lambda, the parameter estimation process approximates the steepest-descent method of optimization. While the latter method is inefficient and slow if used for the entirety of the optimization process, it often helps in getting the process started, especially if initial parameter estimates are poor. | |||
*''Marquardt Lambda adjustment constant'' (RLAMFAC) | |||
*:A real variable, and the factor by which the Marquardt lambda is adjusted. RLAMFAC must be greater than "1.0". When PEST reduces lambda, it divides by RLAMFAC; when it increases lambda, it multiplies by RLAMFAC. PEST reduces lambda if it can. However, if the normal matrix is not positive definite, or if a reduction in lambda does not lower the objective function, PEST has no choice but to increase lambda. | |||
*''Number of lanbdas tested'' (NUMLAM) | |||
*:This integer variable places an upper limit on the number of lambdas that PEST can test during any one optimization iteration. It should normally be set between "5" and "10". For cases where parameters are being adjusted near their upper or lower limits, and for which some parameters are consequently being frozen (thus reducing the dimension of the problem in parameter space), a value closer to "10" may be more appropriate than one closer to "5". This gives PEST a greater chance of adjusting to the reduced problem dimension as parameters are frozen. | |||
*''PHIRATSUF'' | |||
*:A real variable. Stands for "phi ratio sufficient". A value of "0.3" is often appropriate. If it is set too low, model runs may be wasted in search of an objective function reduction which is not possible to achieve, given the linear approximation upon which the optimization equations are based. If it is set too high, PEST may not be given the opportunity of refining lambda in order that its value continues to be optimal as the parameter estimation process progresses. | |||
*:During any one optimization iteration, PEST may calculate a parameter upgrade vector using a number of different Marquardt lambdas. First, it lowers lambda. If this is unsuccessful in lowering the objective function, it then raises lambda. At any stage, if it calculates an objective function which is a fraction PHIRATSUF or less of the starting objective function for that iteration, PEST considers the goal of the current iteration to have been achieved and moves on to the next optimization iteration. | |||
*''PHIREDLAM'' | |||
*:A real variable. A suitable value for PHIREDLAM is often around "0.01". If it is set too large, the criterion for moving on to the next optimization iteration is too easily met, and PEST is not given the opportunity of adjusting lambda to its optimal value for that particular stage of the PEST process. On the other hand, if PHIREDLAM is set too low, PEST will test too many Marquardt lambdas on each optimization iteration when it would be better off starting on a new iteration. | |||
*:If a new/old objective function ratio of PHIRATSUF or less is not achieved, as the effectiveness of different Marquardt lambdas in lowering the objective function is tested, PEST must use some other criterion in deciding when it should move on to the next optimization iteration. This criterion is partly provided by PHIREDLAM. The first lambda that PEST employs in calculating the parameter upgrade vector during any one optimization iteration is the lambda inherited from the previous iteration, possibly reduced by a factor of RLAMFAC (unless it is the first iteration, in which case RLAMBDA1 is used). PEST will try another lambda unless PHIREDLAM reduces the objective function to less than PHIRATSUF of its value at the beginning of the iteration. | |||
*:If the objective function is lower than for the first lambda (and still above PHIRATSUF of the starting objective function), PEST reduces lambda again. Otherwise, it increases lambda to a value greater by a factor of RLAMFAC than the first lambda for the iteration. When attempting to find a more effective lambda by lowering or raising lambda in this fashion, if the objective function begins to rise, PEST accepts the lambda and the corresponding parameter set. This gives rise to the lowest objective function for that iteration and moves on to the next iteration. If the relative reduction in the objective function between the use of two consecutive lambdas is less than PHIREDLAM, PEST determines it is more efficient to begin the next optimization iteration than to continue testing the effect of new Marquardt lambdas. | |||
*''PHIREDSTP'' | |||
*:A real variable used to tell PEST that the optimization process is at an end. For many cases, "0.01" is a suitable value. | |||
*''NPHISTP'' | |||
*:An integer variable used to tell PEST that the optimization process is at an end. For many cases, "4" is a suitable value. Be careful to not set NPHISTP too low if the optimal values for some parameters are near or at their upper or lower bounds. This can cause the magnitude of the parameter upgrade vector to be curtailed over one or a number of optimization iterations to ensure that no parameter value overshoots its bounds. The result may then be smaller reductions in the objective function than would otherwise occur, which may mistakenly appear to be the onset of parameter convergence to the optimal set. | |||
*''NPHINORED'' | |||
*:An integer variable. A value of "3" or "4" is often suitable. If PEST has failed to lower the objective function over NPHINORED successive iterations, it will terminate execution. | |||
*''RELPARSTP'' | |||
*:A real variable. A value of "0.01" is often suitable. If the magnitude of the maximum relative parameter change between optimization iterations is less than RELPARSTP over NRELPAR successive iterations, PEST will terminate. PEST evaluates this change for all adjustable parameters at the end of each optimization iteration and determines the relative parameter change with the highest magnitude. If this maximum relative change is less than RELPARSTP, a counter is advanced by one. If it is greater than RELPARSTP, the counter is zeroed. | |||
*''NRELPAR'' | |||
*:An integer variable. A value of "2" or "3" is often suitable. | |||
==Multistart Parameters dialog== | |||
[[Image:GSSHA Multistart Parameters dialog.png|thumb|right|300 px|''Multistart Parameters'' dialog.]] | |||
The ''Multistart Parameters'' dialog has the following options: | |||
*''Random number seed'' – An integer used to initialize the random number generator. The random number generator's number sequence is determined by this seed. Therefore, if the same seed is used in additional runs, it will produce the same sequence of numbers. | |||
*''# of LM/SLM local searches'' – The number of LM/SLM local searches. A value of "3" is the default. | |||
{{-}} | |||
==Trajectory Repulsion (TR) Parameters dialog== | |||
[[Image:GSSHA Trajectory Repulsion (TR) Parameters.png|thumb|right|300 px|''Trajectory Repulsion (TR) Parameters'' dialog.]] | |||
The ''Trajectory Repulsion (TR) Parameters'' dialog has the following options: | |||
*''Write random number seed file'' – Turn this on to export a random number seed file. | |||
*''Initial random number seed'' – An integer used to initialize the random number generator. The random number generator's number sequence is determined by this seed. Therefore, if the same seed is used in additional runs, it will produce the same sequence of numbers. | |||
*''# of initial function evaluations'' – An integer. The initial number of times to perform function evaluations. | |||
*''# of local searches'' – An integer. The number of local searches. A value of "3" is the default. | |||
*''# of local searches with no objective function improvement'' – An integer. The number of local searches to allow when no objective function improvement is obtained. A value of "2" is the default. | |||
*''Negligible objective function improvement fraction'' – A decimal fraction indicating the amount below which a function improvement is considered negligible. | |||
{{-}} | |||
==Multilevel Single Linkage (MLSL) Parameters dialog== | |||
[[Image:EditMlslParameters.png|thumb|right|300 px|''Multilevel Single Linkage (MLSL) Parameters'' dialog.]] | |||
The ''Multilevel Single Linkage (MLSL) Parameters'' dialog has the following options: | |||
*''MLSL parameter N'' – | |||
*''MLSL parameter gamma'' – | |||
*''MLSL parameter sigma'' – | |||
*''MLSL parameter d2'' – | |||
*''Write random number seed file'' – Turn this on to export a random number seed file. | |||
*''Initial random number seed'' – An integer. An integer used to initialize the random number generator. The random number generator's number sequence is determined by this seed. Therefore, if the same seed is used in additional runs, it will produce the same sequence of numbers. | |||
*''# of MLSL iterations'' – An integer. The number of MLSL iterations to include in the GSSHA run. A value of "50" is the default. | |||
*''# of local searches'' – An integer. The number of local searches. A value of "3" is the default. | |||
*''# of local searches with no objective function improvement'' – An integer. The number of local searches to allow when no objective function improvement is obtained. A value of "4" is the default. | |||
*''# of MLSL iterations with no objective function improvement'' – An integer. The number of MLSL iterations to allow when no objective function improvement is obtained. A value of "5" is the default. | |||
*''Negligible objective function improvement fraction'' – A decimal fraction indicating the amount below which a function improvement is considered negligible. | |||
{{-}} | |||
{{GSSHA Wiki}} | |||
{{WMSMain}} | |||
{{ | |||
}} | |||
[[Category:GSSHA|C]] | [[Category:GSSHA|C]] | ||
[[Category: | [[Category:GSSHA Dialogs|Cal]] | ||
[[Category:Calibration Dialogs]] | |||
[[Category:Calibration]] | |||
Latest revision as of 22:49, 7 July 2020
The GSSHA model allows for both automated and manual calibration. There is a WMS tutorial that describes how to set up a basic calibration model.
GSSHA has a PEST-based automated calibration routine, as described on the GSSHA wiki. WMS writes the parameters, observed data, and calibration files necessary to run GSSHA in any of the supported GSSHA calibration modes. Enter in the needed data by turning on Calibrate in the list on the right in the Job Control dialog.
Clicking on the Edit parameters button brings up the Parameters dialog (below) where parameters can be selected to be calibrated and edited, including the maximum, minimum, and initial values for each parameter.
Defining Key Values
Select adjustable parameters by defining key values as negative integers in the regular GSSHA interface and then assigning starting, minimum, and maximum values to each of these parameters in the Parameters dialog. For example, to set overland Manning’s roughness values as an adjustable model parameter, specify negative integers for the values in the mapping table dialog as shown below:
Then define the key values in the calibration Parameters dialog. The Initialize Parameters from Model button makes defining key values easy. This button searches the model for any parameters that are defined as negative integers and assigns these parameters to the dialog. Starting, minimum, and maximum values should be defined for each key value for the calibration engine. Other parameter information should also be defined, including:
- Whether parameters are fixed or tied to other parameters
- The methods used to calculate derivatives for each parameter
- Defining regularization parameters for prior calibration run information
- Defining preferred values (where a specific value is set as an optimal value for the calibration parameter) or homogeneous values (where one or more calibration parameters are linked to another calibration parameter in an attempt to get these values as close as possible).
Defining Observed Data
Clicking the Observed Data... button in the Calibration Parameters dialog opens the GSSHA Observations dialog (right). In this dialog, enter in various types of observed data (including time series hydrograph data) for each event of the simulation. It is not necessary to associate a rainfall event with each observed data time series.
This dialog shows all the feature points with observed data and allows turning automated calibration on or off for each of these observations. All automated calibration methods in WMS support automated calibration of the following data types at any computation point in the watershed model:
- Overland Depth
- Infiltration Depth
- Surface Moisture
- Grid Suspended Sediment (TSS) Concentration
- Channel Depth
- Channel Flow
- Channel Total Suspended Sediment (TSS) Concentration
- Groundwater Head
- Outlet Hydrograph (only at the watershed outlet point)
- Snow Water Equivalent
- Tile Drain Discharge (in a GSSHA Storm Drain coverage)
The GSSHA Observations dialog can be accessed from the feature point/node attribute dialog in the GSSHA and the GSSHA Storm Drain coverages. If WMS is running the default LM/SLM-based calibration, associate weights with each observation value in the XY series by clicking on the Weights button in the observation window and defining weights for each value. WMS defaults all the weights for observed data points to 1.0, but this value could be modified to give higher weights to certain observed values.
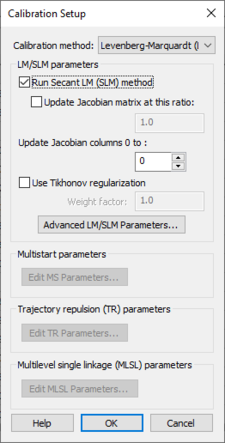
Defining Calibration Setup Parameters
The calibration setup parameters are accessed by clicking the Calibration Setup... button to bring up the Calibration Setup dialog:
The parameters in this dialog are used for creating the calibration control file. This file, along with the parameters and observed data files are written out by WMS when saving a GSSHA project.
The following options are in this dialog:
- Calibration method – A drop-down with a list of calibration methods. The GSSHA wiki has more information on these methods.
- "Levenberg-Marquardt (LM)/Secant LM (SLM)" – Uses local search to optimize model parameters.
- "Multistart (MS)" – Uses global search to optimize model parameters.
- "Trajectory Repulsion (TR)" – Uses global search to optimize model parameters.
- "Multilevel Single Linkage (MLSL)" – Uses global search to optimize model parameters.
- LM/SLM parameters
- Run Secant LM (SLM) method – Turning on this option to run the SLM method sets the input file flag to run the SLM method instead of the LM (Levenberg-Marquardt) local search method. The SLM method is an efficiency enhancement to the LM method and is the default local search optimization method for a GSSHA calibration in the WMS interface.
- Update Jacobian matrix at this ratio – Allows adjusting of the Jacobian matrix update ratio.
- Update Jacobian columns 0 to – Change the number of Jacobian columns.
- Use Tikhonov regularization – Turns Tikhonov regularization off or on. Tikhonov regularization provides a way to adjust the balance between fitting the solution to the observed data and fitting to the regularization relationships by defining a regularization weight factor and running Tikhonov regularization using this weight factor. The GSSHA wiki has more information.
- Advanced LM/SLM Parameters... – Brings up the Advanced LM/SLM Parameters dialog.
- Run Secant LM (SLM) method – Turning on this option to run the SLM method sets the input file flag to run the SLM method instead of the LM (Levenberg-Marquardt) local search method. The SLM method is an efficiency enhancement to the LM method and is the default local search optimization method for a GSSHA calibration in the WMS interface.
- Multistart parameters
- Edit MS Parameters... – Brings up the Multistart Parameters dialog.
- Trajectory repulsion (TR) parameters
- Edit TR Parameters... – Brings up the Trajectory Repulsion (TR) Parameters dialog.
- Multilevel single linkage (MLSL) parameters
- Edit MLSL Parameters... – Brings up the Multilevel Single Linkage (MLSL) Parameters dialog.
- SCE parameters section
- SCE is deprecated in GSSHA and is not supported beginning in WMS 10.0.
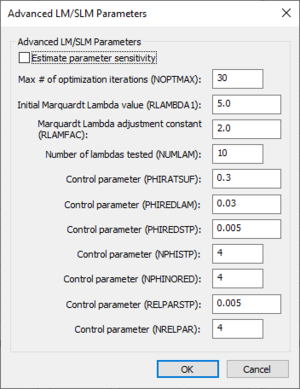
Advanced LM/SLM Parameters dialog
The Advanced LM/SLM Parameters dialog can be used to set advanced calibration settings when running an LM/SLM calibration. Some of the descriptions below were taken from the 2013 PEST User Manual. The following options are available in the Advanced LM/SLM Parameters section:
- Estimate parameter sensitivity
- Turn this option on to write a "-1" to the NOPTMAX parameter in the PEST control file. This tells PEST to only run a sensitivity analysis instead of a full parameter optimization run. The sensitivity file is imported into WMS if reading the calibration solution.
- Max # of optimization iterations (NOPTMAX)
- A value of "20" to "30" is often appropriate. Sets the maximum number of optimization iterations that PEST is permitted to undertake on a particular parameter estimation run. To trigger PEST termination by other criteria more indicative of parameter convergence to an optimal set or of the futility of further processing, set this variable very high.
- If NOPTMAX is set to zero, PEST will not calculate the Jacobian matrix. Instead, it will terminate execution after just one model run. This setting can thus be used when calculating the objective function corresponding to a particular parameter set and/or to inspect observation residuals corresponding to that parameter set.
- Initial Marquardt Lambda value (RLAMBDA1)
- This real variable is the initial Marquardt lambda. An initial value of "1.0" to "10.0" is appropriate for most models. PEST attempts parameter improvement using a number of different Marquardt lambdas during any one optimization iteration. However, in the course of the overall parameter estimation process, the Marquardt lambda generally gets smaller.
- Provide a higher initial Marquardt lambda if PEST complains that the normal matrix is not positive definite. For high values of the Marquardt lambda, the parameter estimation process approximates the steepest-descent method of optimization. While the latter method is inefficient and slow if used for the entirety of the optimization process, it often helps in getting the process started, especially if initial parameter estimates are poor.
- Marquardt Lambda adjustment constant (RLAMFAC)
- A real variable, and the factor by which the Marquardt lambda is adjusted. RLAMFAC must be greater than "1.0". When PEST reduces lambda, it divides by RLAMFAC; when it increases lambda, it multiplies by RLAMFAC. PEST reduces lambda if it can. However, if the normal matrix is not positive definite, or if a reduction in lambda does not lower the objective function, PEST has no choice but to increase lambda.
- Number of lanbdas tested (NUMLAM)
- This integer variable places an upper limit on the number of lambdas that PEST can test during any one optimization iteration. It should normally be set between "5" and "10". For cases where parameters are being adjusted near their upper or lower limits, and for which some parameters are consequently being frozen (thus reducing the dimension of the problem in parameter space), a value closer to "10" may be more appropriate than one closer to "5". This gives PEST a greater chance of adjusting to the reduced problem dimension as parameters are frozen.
- PHIRATSUF
- A real variable. Stands for "phi ratio sufficient". A value of "0.3" is often appropriate. If it is set too low, model runs may be wasted in search of an objective function reduction which is not possible to achieve, given the linear approximation upon which the optimization equations are based. If it is set too high, PEST may not be given the opportunity of refining lambda in order that its value continues to be optimal as the parameter estimation process progresses.
- During any one optimization iteration, PEST may calculate a parameter upgrade vector using a number of different Marquardt lambdas. First, it lowers lambda. If this is unsuccessful in lowering the objective function, it then raises lambda. At any stage, if it calculates an objective function which is a fraction PHIRATSUF or less of the starting objective function for that iteration, PEST considers the goal of the current iteration to have been achieved and moves on to the next optimization iteration.
- PHIREDLAM
- A real variable. A suitable value for PHIREDLAM is often around "0.01". If it is set too large, the criterion for moving on to the next optimization iteration is too easily met, and PEST is not given the opportunity of adjusting lambda to its optimal value for that particular stage of the PEST process. On the other hand, if PHIREDLAM is set too low, PEST will test too many Marquardt lambdas on each optimization iteration when it would be better off starting on a new iteration.
- If a new/old objective function ratio of PHIRATSUF or less is not achieved, as the effectiveness of different Marquardt lambdas in lowering the objective function is tested, PEST must use some other criterion in deciding when it should move on to the next optimization iteration. This criterion is partly provided by PHIREDLAM. The first lambda that PEST employs in calculating the parameter upgrade vector during any one optimization iteration is the lambda inherited from the previous iteration, possibly reduced by a factor of RLAMFAC (unless it is the first iteration, in which case RLAMBDA1 is used). PEST will try another lambda unless PHIREDLAM reduces the objective function to less than PHIRATSUF of its value at the beginning of the iteration.
- If the objective function is lower than for the first lambda (and still above PHIRATSUF of the starting objective function), PEST reduces lambda again. Otherwise, it increases lambda to a value greater by a factor of RLAMFAC than the first lambda for the iteration. When attempting to find a more effective lambda by lowering or raising lambda in this fashion, if the objective function begins to rise, PEST accepts the lambda and the corresponding parameter set. This gives rise to the lowest objective function for that iteration and moves on to the next iteration. If the relative reduction in the objective function between the use of two consecutive lambdas is less than PHIREDLAM, PEST determines it is more efficient to begin the next optimization iteration than to continue testing the effect of new Marquardt lambdas.
- PHIREDSTP
- A real variable used to tell PEST that the optimization process is at an end. For many cases, "0.01" is a suitable value.
- NPHISTP
- An integer variable used to tell PEST that the optimization process is at an end. For many cases, "4" is a suitable value. Be careful to not set NPHISTP too low if the optimal values for some parameters are near or at their upper or lower bounds. This can cause the magnitude of the parameter upgrade vector to be curtailed over one or a number of optimization iterations to ensure that no parameter value overshoots its bounds. The result may then be smaller reductions in the objective function than would otherwise occur, which may mistakenly appear to be the onset of parameter convergence to the optimal set.
- NPHINORED
- An integer variable. A value of "3" or "4" is often suitable. If PEST has failed to lower the objective function over NPHINORED successive iterations, it will terminate execution.
- RELPARSTP
- A real variable. A value of "0.01" is often suitable. If the magnitude of the maximum relative parameter change between optimization iterations is less than RELPARSTP over NRELPAR successive iterations, PEST will terminate. PEST evaluates this change for all adjustable parameters at the end of each optimization iteration and determines the relative parameter change with the highest magnitude. If this maximum relative change is less than RELPARSTP, a counter is advanced by one. If it is greater than RELPARSTP, the counter is zeroed.
- NRELPAR
- An integer variable. A value of "2" or "3" is often suitable.
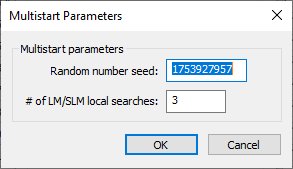
Multistart Parameters dialog
The Multistart Parameters dialog has the following options:
- Random number seed – An integer used to initialize the random number generator. The random number generator's number sequence is determined by this seed. Therefore, if the same seed is used in additional runs, it will produce the same sequence of numbers.
- # of LM/SLM local searches – The number of LM/SLM local searches. A value of "3" is the default.
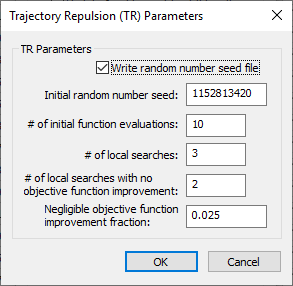
Trajectory Repulsion (TR) Parameters dialog
The Trajectory Repulsion (TR) Parameters dialog has the following options:
- Write random number seed file – Turn this on to export a random number seed file.
- Initial random number seed – An integer used to initialize the random number generator. The random number generator's number sequence is determined by this seed. Therefore, if the same seed is used in additional runs, it will produce the same sequence of numbers.
- # of initial function evaluations – An integer. The initial number of times to perform function evaluations.
- # of local searches – An integer. The number of local searches. A value of "3" is the default.
- # of local searches with no objective function improvement – An integer. The number of local searches to allow when no objective function improvement is obtained. A value of "2" is the default.
- Negligible objective function improvement fraction – A decimal fraction indicating the amount below which a function improvement is considered negligible.
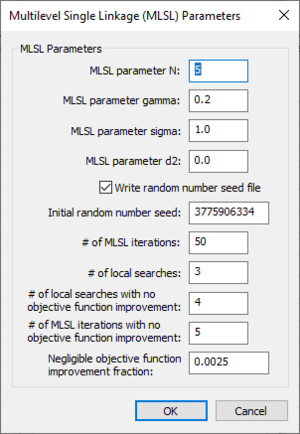
Multilevel Single Linkage (MLSL) Parameters dialog
The Multilevel Single Linkage (MLSL) Parameters dialog has the following options:
- MLSL parameter N –
- MLSL parameter gamma –
- MLSL parameter sigma –
- MLSL parameter d2 –
- Write random number seed file – Turn this on to export a random number seed file.
- Initial random number seed – An integer. An integer used to initialize the random number generator. The random number generator's number sequence is determined by this seed. Therefore, if the same seed is used in additional runs, it will produce the same sequence of numbers.
- # of MLSL iterations – An integer. The number of MLSL iterations to include in the GSSHA run. A value of "50" is the default.
- # of local searches – An integer. The number of local searches. A value of "3" is the default.
- # of local searches with no objective function improvement – An integer. The number of local searches to allow when no objective function improvement is obtained. A value of "4" is the default.
- # of MLSL iterations with no objective function improvement – An integer. The number of MLSL iterations to allow when no objective function improvement is obtained. A value of "5" is the default.
- Negligible objective function improvement fraction – A decimal fraction indicating the amount below which a function improvement is considered negligible.
GSSHA | |
|---|---|
| XMS Wiki Links | Calibration (Automated • Manual • Output) • Channel Routing • Contaminants • Digital Dams • Embankment Arcs • Feature Objects (Arcs • Nodes • Polygons) • File Types • Groundwater • Groups • Hydraulic Structures • Job Control • Join SSURGO Data • Mapping Tables • Maps • Menu • Model Linkage • Multiple Simulations • Nutrients • Observations • Output Control • Overland Soil Erosion • Pipe and Node Parameters • Precipitation • Radar Rainfall • Save GSSHA Project File • Smooth GSSHA Streams • Snowmelt • Solution (Analysis • Data) |
| Related Tools | MWBM Wizard • Using Soil Type Data with GSSHA |
| GSSHA Wiki External Links | GSSHA Wiki: Overview • Primer • User's Manual • Tutorials |
WMS – Watershed Modeling System | ||
|---|---|---|
| Modules: | Terrain Data • Drainage • Map • Hydrologic Modeling • River • GIS • 2D Grid • 2D Scatter |  |
| Models: | CE-QUAL-W2 • GSSHA • HEC-1 • HEC-HMS • HEC-RAS • HSPF • MODRAT • NSS • OC Hydrograph • OC Rational • Rational • River Tools • Storm Drain • SMPDBK • SWMM • TR-20 • TR-55 | |
| Toolbars: | Modules • Macros • Units • Digitize • Static Tools • Dynamic Tools • Drawing • Get Data Tools | |
| Aquaveo | ||